Temperature Salinity Diagram¶
This notebook shows how to plot a temperature-salinity diagram which is weighted by volume using xhistogram.
Output from both MOM5 or MOM6 can be used.
Requirements: The conda/analysis3 (or later) module on ARE. A session with 4 cores is sufficient for this example but more cores will be needed for larger datasets.
Firstly, we load all required modules and start a client.
[1]:
%matplotlib inline
import xarray as xr
import numpy as np
import cosima_cookbook as cc
from dask.distributed import Client
import gsw
from xhistogram.xarray import histogram as xhistogram
import cf_xarray as cfxr
import warnings
warnings.simplefilter('ignore')
## plotting
import matplotlib.pyplot as plt
import cmocean.cm as cmo
import matplotlib.colors as colors
[2]:
client = Client()
client
[2]:
Client
Client-646264f3-a062-11ed-a4cd-00000772fe80
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: /proxy/39221/status |
Cluster Info
LocalCluster
817f5334
| Dashboard: /proxy/39221/status | Workers: 7 |
| Total threads: 28 | Total memory: 112.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-3c8dbc89-bb9d-4941-9ec9-6ddc23628db0
| Comm: tcp://127.0.0.1:46113 | Workers: 7 |
| Dashboard: /proxy/39221/status | Total threads: 28 |
| Started: Just now | Total memory: 112.00 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:45779 | Total threads: 4 |
| Dashboard: /proxy/40195/status | Memory: 16.00 GiB |
| Nanny: tcp://127.0.0.1:33593 | |
| Local directory: /jobfs/71077001.gadi-pbs/dask-worker-space/worker-3qr7ppqe | |
Worker: 1
| Comm: tcp://127.0.0.1:37439 | Total threads: 4 |
| Dashboard: /proxy/37985/status | Memory: 16.00 GiB |
| Nanny: tcp://127.0.0.1:42647 | |
| Local directory: /jobfs/71077001.gadi-pbs/dask-worker-space/worker-0cget0n8 | |
Worker: 2
| Comm: tcp://127.0.0.1:36971 | Total threads: 4 |
| Dashboard: /proxy/45137/status | Memory: 16.00 GiB |
| Nanny: tcp://127.0.0.1:44953 | |
| Local directory: /jobfs/71077001.gadi-pbs/dask-worker-space/worker-hopukhfh | |
Worker: 3
| Comm: tcp://127.0.0.1:38489 | Total threads: 4 |
| Dashboard: /proxy/32955/status | Memory: 16.00 GiB |
| Nanny: tcp://127.0.0.1:44723 | |
| Local directory: /jobfs/71077001.gadi-pbs/dask-worker-space/worker-befoobp0 | |
Worker: 4
| Comm: tcp://127.0.0.1:46429 | Total threads: 4 |
| Dashboard: /proxy/35921/status | Memory: 16.00 GiB |
| Nanny: tcp://127.0.0.1:43179 | |
| Local directory: /jobfs/71077001.gadi-pbs/dask-worker-space/worker-o9u5k7k2 | |
Worker: 5
| Comm: tcp://127.0.0.1:44453 | Total threads: 4 |
| Dashboard: /proxy/38357/status | Memory: 16.00 GiB |
| Nanny: tcp://127.0.0.1:39165 | |
| Local directory: /jobfs/71077001.gadi-pbs/dask-worker-space/worker-sb4zzmr5 | |
Worker: 6
| Comm: tcp://127.0.0.1:33031 | Total threads: 4 |
| Dashboard: /proxy/40619/status | Memory: 16.00 GiB |
| Nanny: tcp://127.0.0.1:40605 | |
| Local directory: /jobfs/71077001.gadi-pbs/dask-worker-space/worker-ob69v54g | |
Load data¶
Select output from MOM5 or MOM6 and coordinates of meridional section.
[3]:
lon = -25 # longitude of meridional section
lat = slice(-90, -37) # latitude range of section
session = cc.database.create_session()
Next, choose an experiment of any resolution. Here, only 1 year is selected; if you want to use a longer time period, you might need more resources!
[4]:
# dictionary of experiment name and time for mom5 and mom6
expt_args = {
"mom5": {
"expt": "01deg_jra55v13_ryf9091",
"start_time": "1991-01-01",
"end_time": "1991-12-31"},
"mom6": {
"expt": "panant-01-zstar-v13",
"start_time": "1991-01-01",
"end_time": "1991-12-31"}}
[5]:
# dictionary of variable names and time for mom5 and mom6
vars_args = {
"mom5": {
"var_temp": 'temp',
"var_salt": 'salt',
"var_area": 'area_t'},
"mom6": {
"var_temp": 'thetao',
"var_salt": 'so',
"var_area": 'areacello'}}
We define below a few functions used to load output (temperature or salinity) and compute the histogram for the T-S diagram.
[6]:
def gsw_SA_from_SP(salt, lon_name):
"""function to convert practical salinity to absolute salinity
using the Gibbs SeaWater (GSW) Oceanographic Toolbox of TEOS-10
(https://teos-10.github.io/GSW-Python/)
input:
-salt: practical salinity (array)
-lon_name: name of the longitude (str)
output:
-salt_abs: absolute salinity (array)
"""
pres = gsw.p_from_z(-salt.cf['vertical'], salt.cf['latitude'])
salt_abs = gsw.SA_from_SP(salt, pres, salt[lon_name], salt.cf['latitude'])
salt_abs.attrs = {'units': 'Absolute Salinity (g/kg)'}
return salt_abs
[7]:
# a function to load salinity
def load_salinity(model):
salt = cc.querying.getvar(
variable=vars_args[model].get('var_salt'), session=session, frequency='1 monthly',
**expt_args[model])
lon_name = salt.cf['longitude'].name
# select longitude, latitude range and time
salt = salt.cf.sel(longitude=lon, method='nearest').cf.sel(
latitude=lat, time=slice(expt_args[model].get('start_time'),
expt_args[model].get('end_time')))
# convert from practical to absolute salinity
salt = gsw_SA_from_SP(salt, lon_name)
return salt
[8]:
# a function to load temperature
def load_temperature(model):
temp = cc.querying.getvar(
variable=vars_args[model].get('var_temp'), session=session, frequency='1 monthly',
**expt_args[model])
# select longitude, latitude range and time
temp = temp.cf.sel(longitude=lon, method='nearest').cf.sel(
latitude=lat, time=slice(expt_args[model].get('start_time'),
expt_args[model].get('end_time')))
if model == 'mom5':
temp = temp - 273.15
elif model == 'mom6':
# convert potential to conservative temperature
temp = gsw.conversions.CT_from_pt(salt, temp)
temp.name = 'temp'
temp.attrs = {'units': 'Conservative temperature (°C)'}
return temp
[9]:
# a function to load area of grid cells
def load_grid_areas(model):
area = cc.querying.getvar(
variable=vars_args[model].get('var_area'), session=session, n=1,
**expt_args[model])
if model == 'mom5':
area = area.drop(['geolon_t', 'geolat_t'])
# select longitude and latitude range
area = area.cf.sel(longitude=lon, method='nearest').cf.sel(
latitude=lat)
return area
[10]:
# a function that computes the temperature and salinity bins for 2D histogram
def compute_TS_bins(salt, temp, area):
temp_bins = np.arange(np.floor(temp.min().values), np.ceil(temp.max().values), 0.5)
salt_bins = np.arange(np.floor(salt.min().values), np.ceil(salt.max().values), 0.1)
# for density contours in TS diagram
temp_bins_mesh,salt_bins_mesh = np.meshgrid(temp_bins, salt_bins)
TS_density = gsw.density.sigma2(salt_bins_mesh, temp_bins_mesh)
# volume of grid cells to account for varying grid cells especially in the vertical
vol = (temp.cf['vertical'] * area)
# 2D histogram of temperature and salinity weighted by volume
TS_hist = xhistogram(
temp, salt, bins=(temp_bins, salt_bins), weights=vol)
TS_hist = TS_hist.where(TS_hist != 0).compute()
return TS_hist, TS_density, salt_bins_mesh, temp_bins_mesh
Now let’s select a model, load everything, and compute the TS diagram
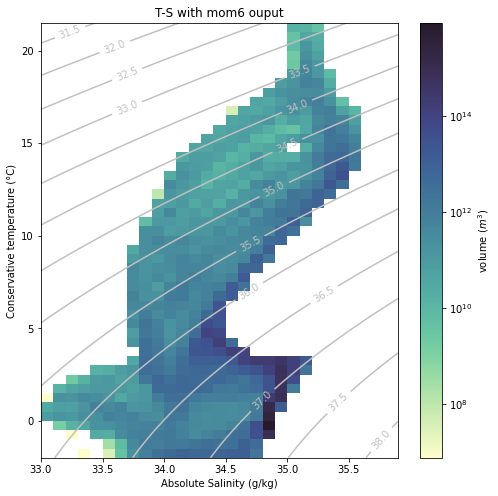
MOM5¶
[11]:
model = 'mom5' # 'mom5' or 'mom6'
salt = load_salinity(model)
temp = load_temperature(model)
area = load_grid_areas(model)
Calculate and plot the TS diagram¶
[12]:
TS_hist, TS_density, salt_bins_mesh, temp_bins_mesh = compute_TS_bins(salt, temp, area)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
[13]:
plt.figure(figsize=(8, 8))
# logarithmic colormap to show both TS bins with smaller and
# larger volumes (e.g. NADW, AABW)
norm=colors.LogNorm(
vmin=TS_hist.min().values, vmax=TS_hist.max().values)
# volume weighted histogram of T and S
TS_hist.plot(cmap=cmo.deep, norm=norm,
cbar_kwargs=dict(label='volume ($m^{3}$)'))
# density contours
cs = plt.contour(
salt_bins_mesh, temp_bins_mesh, TS_density, colors='silver',
levels=np.arange(np.floor(TS_density.min()),
np.ceil(TS_density.max()), .5))
plt.clabel(cs, inline=True)
plt.xlabel(salt.units)
plt.ylabel(temp.units)
plt.title("T-S with "+model+" ouput");
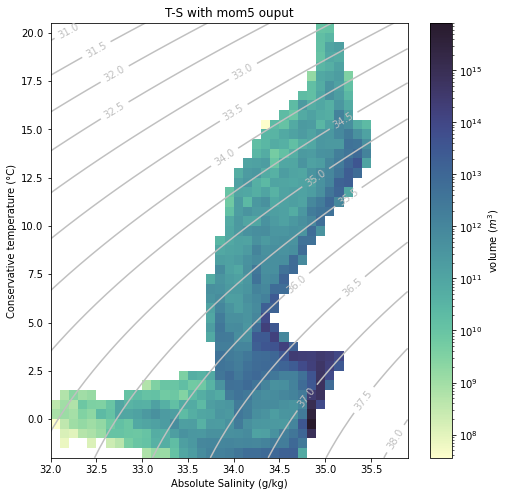
MOM6¶
Now let’s do the same but selecting mom6 as our model.
[14]:
model = 'mom6' # 'mom5' or 'mom6'
salt = load_salinity(model)
temp = load_temperature(model)
area = load_grid_areas(model)
TS_hist, TS_density, salt_bins_mesh, temp_bins_mesh = compute_TS_bins(salt, temp, area)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:611: RuntimeWarning: All-NaN slice encountered
return np.nanmin(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
/g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/dask/array/reductions.py:640: RuntimeWarning: All-NaN slice encountered
return np.nanmax(x_chunk, axis=axis, keepdims=keepdims)
[15]:
plt.figure(figsize=(8, 8))
# logarithmic colormap to show both TS bins with smaller and
# larger volumes (e.g. NADW, AABW)
norm=colors.LogNorm(
vmin=TS_hist.min().values, vmax=TS_hist.max().values)
# volume weighted histogram of T and S
TS_hist.plot(cmap=cmo.deep, norm=norm,
cbar_kwargs=dict(label='volume ($m^{3}$)'))
# density contours
cs = plt.contour(
salt_bins_mesh, temp_bins_mesh, TS_density, colors='silver',
levels=np.arange(np.floor(TS_density.min()),
np.ceil(TS_density.max()), .5))
plt.clabel(cs, inline=True)
plt.xlabel(salt.units)
plt.ylabel(temp.units)
plt.title("T-S with "+model+" ouput");