Model Agnostic Analysis¶
Introduction¶
In this tutorial model agnostic analysis means writing your notebook so that it can easily be used with any CF compliant data source.
What are the CF Conventions?¶
From CF Metadata conventions:
The CF metadata conventions are designed to promote the processing and sharing of files created with the NetCDF API. The conventions define metadata that provide a definitive description of what the data in each variable represents, and the spatial and temporal properties of the data. This enables users of data from different sources to decide which quantities are comparable, and facilitates building applications with powerful extraction, regridding, and display capabilities. The CF convention includes a standard name table, which defines strings that identify physical quantities.
In most cases the model output data accessed through the COSIMA Cookbook complies with some version of the CF conventions, enough to be usable for model agnostic analysis.
Why bother?¶
Model agnostic means the same code can work for multiple models. This makes your code more usable by you and by others. You no longer need to have different versions of code for different models. It makes you and any one who uses your code more productive. It allows for common tasks to be abstracted into general methods that can be more easily reused, meaning less code needs to be written and maintained. This is an enormous produtivity boost.
How is model agnostic analysis achieved?¶
This can be achieved by using packages that enable this: - cf_xarray for generalised coordinate naming - xgcm to make grid operations generic across data - pint and pint-xarray for handling units easily and robustly
Example¶
Introduction¶
This example uses an example analysis, shows how the this might be done in a traditional, model specific, manner, and then implements the same analysis in a model agnostic way.
First step is to import necessary libaries.
[1]:
%matplotlib inline
import cosima_cookbook as cc
import matplotlib.pyplot as plt
import xarray as xr
import numpy as np
import cf_xarray as cfxr
import pint_xarray
from pint import application_registry as ureg
import cf_xarray.units
import cmocean as cm
import cartopy.crs as ccrs
import cartopy.feature as cft
cf_xarray works best when xarray keeps attributes by default.
[2]:
xr.set_options(keep_attrs=True);
Load a dataset using COSIMA Cookbook, so first open a session to the default database
[3]:
session = cc.database.create_session()
Now load surface temperature data from a 0.25\(^\circ\) global MOM5 model
[4]:
experiment = '025deg_jra55v13_iaf_gmredi6'
variable = 'surface_temp'
SST = cc.querying.getvar(experiment, variable, session, frequency='1 monthly', n=12)
This is a 3D dataset in latitude, longitude and time:
[5]:
SST
[5]:
<xarray.DataArray 'surface_temp' (time: 288, yt_ocean: 1080, xt_ocean: 1440)>
dask.array<concatenate, shape=(288, 1080, 1440), dtype=float32, chunksize=(1, 540, 720), chunktype=numpy.ndarray>
Coordinates:
* xt_ocean (xt_ocean) float64 -279.9 -279.6 -279.4 ... 79.38 79.62 79.88
* yt_ocean (yt_ocean) float64 -81.08 -80.97 -80.87 ... 89.74 89.84 89.95
* time (time) datetime64[ns] 1958-01-14T12:00:00 ... 1981-12-14T12:00:00
Attributes:
long_name: Conservative temperature
units: deg_C
valid_range: [-10. 500.]
cell_methods: time: mean
time_avg_info: average_T1,average_T2,average_DT
coordinates: geolon_t geolat_t
ncfiles: ['/g/data/hh5/tmp/cosima/access-om2-025/025deg_jra55v13_i...Model specific¶
First do this as it might usually be done, in a model specific manner:
Use the time coordinate name in the mean function
Subtract a hard-coded value to convert the temperature degrees celcius from degrees Kelvin (the meta-data says the units are
deg_Cbut this is clearly incorrect)
[6]:
SST_time_mean = SST.mean('time') - 273.15
SST_time_mean
[6]:
<xarray.DataArray 'surface_temp' (yt_ocean: 1080, xt_ocean: 1440)>
dask.array<sub, shape=(1080, 1440), dtype=float32, chunksize=(540, 720), chunktype=numpy.ndarray>
Coordinates:
* xt_ocean (xt_ocean) float64 -279.9 -279.6 -279.4 ... 79.38 79.62 79.88
* yt_ocean (yt_ocean) float64 -81.08 -80.97 -80.87 ... 89.74 89.84 89.95
Attributes:
long_name: Conservative temperature
units: deg_C
valid_range: [-10. 500.]
cell_methods: time: mean
time_avg_info: average_T1,average_T2,average_DT
coordinates: geolon_t geolat_t
ncfiles: ['/g/data/hh5/tmp/cosima/access-om2-025/025deg_jra55v13_i...Now plot the result
[7]:
plt.figure(figsize=(12, 6))
ax = plt.axes(projection=ccrs.Robinson())
SST_time_mean.plot(ax=ax,
x='xt_ocean', y='yt_ocean',
transform=ccrs.PlateCarree(),
vmin=-2, vmax=30, extend='both',
cmap=cm.cm.thermal)
ax.coastlines();
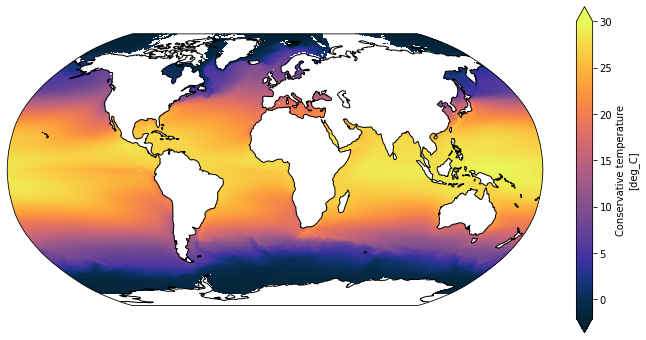
Note that the arctic is not correctly respresented due to the 1D lat/lon coordinates not being correct in the tripole area. See the Making Maps with cartopy Tutorial for more information.
Model agnostic¶
Now do the same calculation in a model agnostic manner
For this data it is necessary to correct the units attribute first. This shouldn’t be necessary if the metadata is correct
[8]:
SST.attrs['units'] = 'K'
Now use pint to ensure this is in degrees C. Note that if the data was originally in degrees celcius this would be fine, it would do nothing. So this is a way of catering for any temperature units that are understood by pint in a transparent way. Note the call to quantify which invokes pint’s machinery to parse the units and allow unit conversions.
[9]:
SST = SST.pint.quantify().pint.to('C')
[10]:
SST
[10]:
<xarray.DataArray 'surface_temp' (time: 288, yt_ocean: 1080, xt_ocean: 1440)>
<Quantity(dask.array<truediv, shape=(288, 1080, 1440), dtype=float32, chunksize=(1, 540, 720), chunktype=numpy.ndarray>, 'degree_Celsius')>
Coordinates:
* xt_ocean (xt_ocean) float64 -279.9 -279.6 -279.4 ... 79.38 79.62 79.88
* yt_ocean (yt_ocean) float64 -81.08 -80.97 -80.87 ... 89.74 89.84 89.95
* time (time) datetime64[ns] 1958-01-14T12:00:00 ... 1981-12-14T12:00:00
Attributes:
long_name: Conservative temperature
valid_range: [-10. 500.]
cell_methods: time: mean
time_avg_info: average_T1,average_T2,average_DT
coordinates: geolon_t geolat_t
ncfiles: ['/g/data/hh5/tmp/cosima/access-om2-025/025deg_jra55v13_i...Now take the time mean, but this time use the cf accessor to automatically determine the name of the time dimension. cf_xarray checks the names of variables and coordinates, and associated metadata to try and infer information about the data based on the CF conventions.
To see what cf_xarray information is available just evaluate the accessor:
[11]:
SST.cf
[11]:
Coordinates:
- CF Axes: * X: ['xt_ocean']
* Y: ['yt_ocean']
* T: ['time']
Z: n/a
- CF Coordinates: * longitude: ['xt_ocean']
* latitude: ['yt_ocean']
* time: ['time']
vertical: n/a
- Cell Measures: area, volume: n/a
- Standard Names: n/a
- Bounds: n/a
In this case it has found X, Y and T axes, and longitude, latitude and time axes. These are now accessible like a dict using the cf accessor. Note that this returns the actual coordinate, and many functions just want a simple string argument, which is the name of the coordinate.
cf_xarray also wraps many standard xarray functions allowing cf names to be used, which are automatically converted to the name in the data.
The upshot: just use the cf accessor and then append the required function and use the standard CF coordinate name (in this case they are the same, time, but that is not guaranteed)
[12]:
SST_time_mean = SST.cf.mean('time')
SST_time_mean
[12]:
<xarray.DataArray 'surface_temp' (yt_ocean: 1080, xt_ocean: 1440)>
<Quantity(dask.array<mean_agg-aggregate, shape=(1080, 1440), dtype=float32, chunksize=(540, 720), chunktype=numpy.ndarray>, 'degree_Celsius')>
Coordinates:
* xt_ocean (xt_ocean) float64 -279.9 -279.6 -279.4 ... 79.38 79.62 79.88
* yt_ocean (yt_ocean) float64 -81.08 -80.97 -80.87 ... 89.74 89.84 89.95
Attributes:
long_name: Conservative temperature
valid_range: [-10. 500.]
cell_methods: time: mean
time_avg_info: average_T1,average_T2,average_DT
coordinates: geolon_t geolat_t
ncfiles: ['/g/data/hh5/tmp/cosima/access-om2-025/025deg_jra55v13_i...Using the cf_xarray wrapped function makes the code more legible and easier to write, i.e.
SST.cf.mean('time')
compared to
SST.mean(SST.cf['time'].name)
In the same way the cf accessor can be used in the plot and the CF names for latitude and longitude used as x and y arguments
[13]:
plt.figure(figsize=(12, 6))
ax = plt.axes(projection=ccrs.Robinson())
SST_time_mean.cf.plot(ax=ax,
x='longitude', y='latitude',
transform=ccrs.PlateCarree(),
vmin=-2, vmax=30, extend='both',
cmap=cm.cm.thermal)
ax.coastlines();
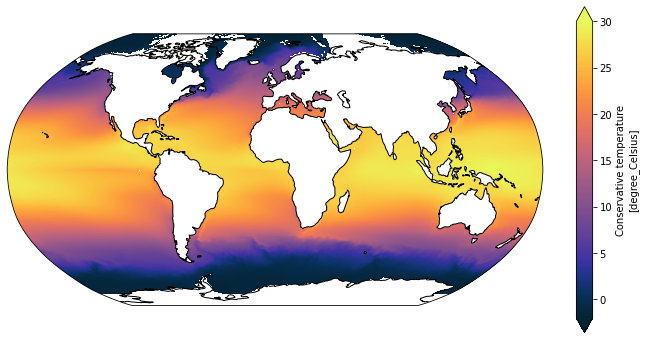
Putting this into practice¶
Above a model agnostic version of some code was demonstrated, but that doesn’t utilise the full power of what it is capable of. The model agnostic code can now be easily turned into a function that accepts an xarray DataArray:
[14]:
def plot_global_temp_in_degrees_celcius(da):
# Take the time mean of da and plot a global temperature field in a Robinson projection
#
# Input DataArray (da) should be a 3D array of latitude, longitude and time.
da = da.pint.quantify().pint.to('C')
da_time_mean = da.cf.mean('time')
plt.figure(figsize=(12, 6))
ax = plt.axes(projection=ccrs.Robinson())
da_time_mean.cf.plot(ax=ax,
x='longitude', y='latitude',
transform=ccrs.PlateCarree(),
vmin=-2, vmax=30, extend='both',
cmap=cm.cm.thermal)
ax.coastlines();
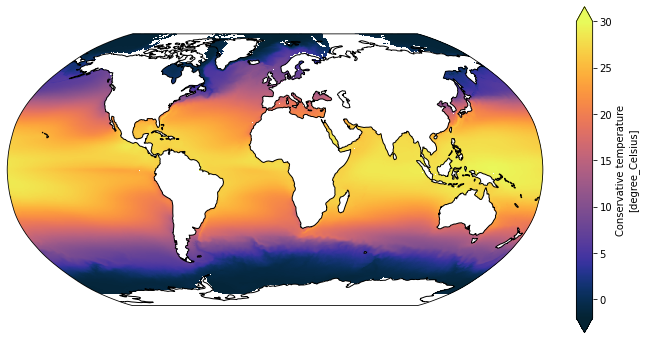
Try it out with the SST data used above
[15]:
plot_global_temp_in_degrees_celcius(SST)
Ok, so now try on the output from a different experiment and model (MOM6):
[16]:
experiment = 'OM4_025.JRA_RYF'
variable = 'tos'
SST_mom6 = cc.querying.getvar(experiment, variable, session, frequency='1 monthly', n=12)
[17]:
SST_mom6
[17]:
<xarray.DataArray 'tos' (time: 144, yh: 1080, xh: 1440)>
dask.array<concatenate, shape=(144, 1080, 1440), dtype=float32, chunksize=(1, 540, 720), chunktype=numpy.ndarray>
Coordinates:
* xh (xh) float64 -299.7 -299.5 -299.2 -299.0 ... 59.53 59.78 60.03
* yh (yh) float64 -80.39 -80.31 -80.23 -80.15 ... 89.73 89.84 89.95
* time (time) object 1900-01-16 12:00:00 ... 1911-12-16 12:00:00
Attributes:
units: degC
long_name: Sea Surface Temperature
cell_methods: area:mean yh:mean xh:mean time: mean
cell_measures: area: areacello
time_avg_info: average_T1,average_T2,average_DT
standard_name: sea_surface_temperature
ncfiles: ['/g/data/ik11/outputs/mom6-om4-025/OM4_025.JRA_RYF/outpu...
contact: Andy Hogg
email: Andy.Hogg@anu.edu.au
created: 2021-11-01
description: 0.25 degree OM4 (MOM6+SIS2) global model configuration un...Check to see it has correctly parsed the CF information. It is not necessary to print this out, but interesting, and note it has quite different index and coordinate names
[18]:
SST_mom6.cf
[18]:
Coordinates:
- CF Axes: * X: ['xh']
* Y: ['yh']
* T: ['time']
Z: n/a
- CF Coordinates: * longitude: ['xh']
* latitude: ['yh']
* time: ['time']
vertical: n/a
- Cell Measures: area, volume: n/a
- Standard Names: n/a
- Bounds: n/a
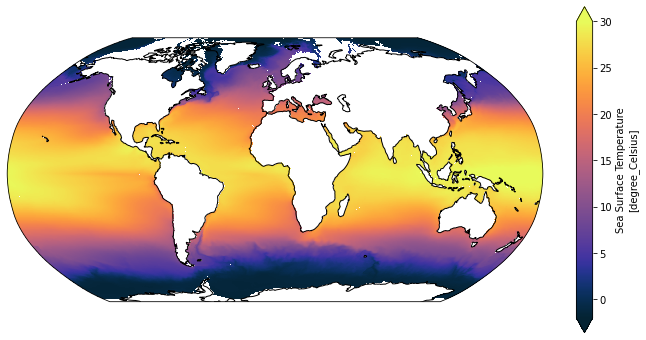
Use the function from above which also worked on MOM5 data with very different coordinates
[19]:
plot_global_temp_in_degrees_celcius(SST_mom6)
What to do when it goes wrong¶
The model agnostic function worked flawlessly with two different ocean data sets, after the units were corrected in the MOM5 data. What about some ice data?
Using the same experiment from which the first SST data was obtained, load the ice air temperature variable
[20]:
experiment = '025deg_jra55v13_iaf_gmredi6'
variable = 'Tair_m'
ice_air_temp = cc.querying.getvar(experiment, variable, session, frequency='1 monthly', n=12)
[21]:
ice_air_temp
[21]:
<xarray.DataArray 'Tair_m' (time: 12, nj: 1080, ni: 1440)>
dask.array<concatenate, shape=(12, 1080, 1440), dtype=float32, chunksize=(1, 1080, 1440), chunktype=numpy.ndarray>
Coordinates:
* time (time) datetime64[ns] 1958-02-01 1958-03-01 ... 1959-01-01
TLON (nj, ni) float32 dask.array<chunksize=(1080, 1440), meta=np.ndarray>
TLAT (nj, ni) float32 dask.array<chunksize=(1080, 1440), meta=np.ndarray>
ULON (nj, ni) float32 dask.array<chunksize=(1080, 1440), meta=np.ndarray>
ULAT (nj, ni) float32 dask.array<chunksize=(1080, 1440), meta=np.ndarray>
Dimensions without coordinates: nj, ni
Attributes:
units: C
long_name: air temperature
cell_measures: area: tarea
cell_methods: time: mean
time_rep: averaged
ncfiles: ['/g/data/hh5/tmp/cosima/access-om2-025/025deg_jra55v13_i...And try the generic routine
[22]:
plot_global_temp_in_degrees_celcius(ice_air_temp)
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[22], line 1
----> 1 plot_global_temp_in_degrees_celcius(ice_air_temp)
Cell In[14], line 12, in plot_global_temp_in_degrees_celcius(da)
9 plt.figure(figsize=(12, 6))
10 ax = plt.axes(projection=ccrs.Robinson())
---> 12 da_time_mean.cf.plot(ax=ax,
13 x='longitude', y='latitude',
14 transform=ccrs.PlateCarree(),
15 vmin=-2, vmax=30, extend='both',
16 cmap=cm.cm.thermal)
18 ax.coastlines()
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:907, in _CFWrappedPlotMethods.__call__(self, *args, **kwargs)
898 """
899 Allows .plot()
900 """
901 plot = _getattr(
902 obj=self._obj,
903 attr="plot",
904 accessor=self.accessor,
905 key_mappers=dict.fromkeys(self._keys, (_single(_get_all),)),
906 )
--> 907 return self._plot_decorator(plot)(*args, **kwargs)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:890, in _CFWrappedPlotMethods._plot_decorator.<locals>._plot_wrapper(*args, **kwargs)
887 kwargs = self._process_x_or_y(kwargs, "y", skip=kwargs.get("x"))
889 # Now set some nice properties
--> 890 kwargs = self._set_axis_props(kwargs, "x")
891 kwargs = self._set_axis_props(kwargs, "y")
893 return func(*args, **kwargs)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:854, in _CFWrappedPlotMethods._set_axis_props(self, kwargs, key)
852 if value:
853 if value in self.accessor.keys():
--> 854 var = self.accessor[value]
855 else:
856 var = self._obj[value]
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:2516, in CFDataArrayAccessor.__getitem__(self, key)
2511 if not isinstance(key, str):
2512 raise KeyError(
2513 f"Cannot use a list of keys with DataArrays. Expected a single string. Received {key!r} instead."
2514 )
-> 2516 return _getitem(self, key)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:668, in _getitem(accessor, key, skip)
666 names = _get_all(obj, k)
667 names = drop_bounds(names)
--> 668 check_results(names, k)
669 successful[k] = bool(names)
670 coords.extend(names)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:647, in _getitem.<locals>.check_results(names, key)
645 def check_results(names, key):
646 if scalar_key and len(names) > 1:
--> 647 raise KeyError(
648 f"Receive multiple variables for key {key!r}: {names}. "
649 f"Expected only one. Please pass a list [{key!r}] "
650 f"instead to get all variables matching {key!r}."
651 )
KeyError: "Receive multiple variables for key 'longitude': ['ULON', 'TLON']. Expected only one. Please pass a list ['longitude'] instead to get all variables matching 'longitude'."

The error message is
"Receive multiple variables for key 'longitude': ['TLON', 'ULON']. Expected only one. Please pass a list ['longitude'] instead to get all variables matching 'longitude'."
This suggests that cf_xarray has found multiple longitude coordinates TLON and ULON and doesn’t know how to resolve this automatically.
Inspecting the cf information doesn’t show multiple axes like it does in the documentation:
[23]:
ice_air_temp.cf
[23]:
Coordinates:
- CF Axes: X, Y, Z, T: n/a
- CF Coordinates: longitude: ['TLON']
latitude: ['TLAT']
vertical, time: n/a
- Cell Measures: area, volume: n/a
- Standard Names: n/a
- Bounds: n/a
This is a bug, taking the mean alters the coordinates:
[24]:
ice_air_temp.cf.mean('time').cf
[24]:
Coordinates:
- CF Axes: X, Y, Z, T: n/a
- CF Coordinates: longitude: ['TLON', 'ULON']
latitude: ['TLAT', 'ULAT']
vertical, time: n/a
- Cell Measures: area, volume: n/a
- Standard Names: n/a
- Bounds: n/a
So the solution is to drop the redundant velocity grid:
[25]:
ice_air_temp = ice_air_temp.drop_vars(['ULON', 'ULAT'])
Now trying to plot again using the generic function and there is another error:
[26]:
plot_global_temp_in_degrees_celcius(ice_air_temp)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[26], line 1
----> 1 plot_global_temp_in_degrees_celcius(ice_air_temp)
Cell In[14], line 12, in plot_global_temp_in_degrees_celcius(da)
9 plt.figure(figsize=(12, 6))
10 ax = plt.axes(projection=ccrs.Robinson())
---> 12 da_time_mean.cf.plot(ax=ax,
13 x='longitude', y='latitude',
14 transform=ccrs.PlateCarree(),
15 vmin=-2, vmax=30, extend='both',
16 cmap=cm.cm.thermal)
18 ax.coastlines()
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:907, in _CFWrappedPlotMethods.__call__(self, *args, **kwargs)
898 """
899 Allows .plot()
900 """
901 plot = _getattr(
902 obj=self._obj,
903 attr="plot",
904 accessor=self.accessor,
905 key_mappers=dict.fromkeys(self._keys, (_single(_get_all),)),
906 )
--> 907 return self._plot_decorator(plot)(*args, **kwargs)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:893, in _CFWrappedPlotMethods._plot_decorator.<locals>._plot_wrapper(*args, **kwargs)
890 kwargs = self._set_axis_props(kwargs, "x")
891 kwargs = self._set_axis_props(kwargs, "y")
--> 893 return func(*args, **kwargs)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cf_xarray/accessor.py:598, in _getattr.<locals>.wrapper(*args, **kwargs)
594 posargs, arguments = accessor._process_signature(
595 func, args, kwargs, key_mappers
596 )
597 final_func = extra_decorator(func) if extra_decorator else func
--> 598 result = final_func(*posargs, **arguments)
599 if wrap_classes and isinstance(result, _WRAPPED_CLASSES):
600 result = _CFWrappedClass(result, accessor)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/xarray/plot/accessor.py:46, in DataArrayPlotAccessor.__call__(self, **kwargs)
44 @functools.wraps(dataarray_plot.plot, assigned=("__doc__", "__annotations__"))
45 def __call__(self, **kwargs) -> Any:
---> 46 return dataarray_plot.plot(self._da, **kwargs)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/xarray/plot/dataarray_plot.py:312, in plot(darray, row, col, col_wrap, ax, hue, subplot_kws, **kwargs)
308 plotfunc = hist
310 kwargs["ax"] = ax
--> 312 return plotfunc(darray, **kwargs)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/xarray/plot/dataarray_plot.py:1613, in _plot2d.<locals>.newplotfunc(***failed resolving arguments***)
1609 raise ValueError("plt.imshow's `aspect` kwarg is not available in xarray")
1611 ax = get_axis(figsize, size, aspect, ax, **subplot_kws)
-> 1613 primitive = plotfunc(
1614 xplt,
1615 yplt,
1616 zval,
1617 ax=ax,
1618 cmap=cmap_params["cmap"],
1619 vmin=cmap_params["vmin"],
1620 vmax=cmap_params["vmax"],
1621 norm=cmap_params["norm"],
1622 **kwargs,
1623 )
1625 # Label the plot with metadata
1626 if add_labels:
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/xarray/plot/dataarray_plot.py:2340, in pcolormesh(x, y, z, ax, xscale, yscale, infer_intervals, **kwargs)
2337 y = _infer_interval_breaks(y, axis=0, scale=yscale)
2339 ax.grid(False)
-> 2340 primitive = ax.pcolormesh(x, y, z, **kwargs)
2342 # by default, pcolormesh picks "round" values for bounds
2343 # this results in ugly looking plots with lots of surrounding whitespace
2344 if not hasattr(ax, "projection") and x.ndim == 1 and y.ndim == 1:
2345 # not a cartopy geoaxis
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cartopy/mpl/geoaxes.py:318, in _add_transform.<locals>.wrapper(self, *args, **kwargs)
313 raise ValueError(f'Invalid transform: Spherical {func.__name__} '
314 'is not supported - consider using '
315 'PlateCarree/RotatedPole.')
317 kwargs['transform'] = transform
--> 318 return func(self, *args, **kwargs)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/cartopy/mpl/geoaxes.py:1797, in GeoAxes.pcolormesh(self, *args, **kwargs)
1794 # Add in an argument checker to handle Matplotlib's potential
1795 # interpolation when coordinate wraps are involved
1796 args, kwargs = self._wrap_args(*args, **kwargs)
-> 1797 result = matplotlib.axes.Axes.pcolormesh(self, *args, **kwargs)
1798 # Wrap the quadrilaterals if necessary
1799 result = self._wrap_quadmesh(result, **kwargs)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/matplotlib/__init__.py:1414, in _preprocess_data.<locals>.inner(ax, data, *args, **kwargs)
1411 @functools.wraps(func)
1412 def inner(ax, *args, data=None, **kwargs):
1413 if data is None:
-> 1414 return func(ax, *map(sanitize_sequence, args), **kwargs)
1416 bound = new_sig.bind(ax, *args, **kwargs)
1417 auto_label = (bound.arguments.get(label_namer)
1418 or bound.kwargs.get(label_namer))
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/matplotlib/axes/_axes.py:6064, in Axes.pcolormesh(self, alpha, norm, cmap, vmin, vmax, shading, antialiased, *args, **kwargs)
6061 shading = shading.lower()
6062 kwargs.setdefault('edgecolors', 'none')
-> 6064 X, Y, C, shading = self._pcolorargs('pcolormesh', *args,
6065 shading=shading, kwargs=kwargs)
6066 coords = np.stack([X, Y], axis=-1)
6067 # convert to one dimensional array
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.07/lib/python3.9/site-packages/matplotlib/axes/_axes.py:5539, in Axes._pcolorargs(self, funcname, shading, *args, **kwargs)
5537 if funcname == 'pcolormesh':
5538 if np.ma.is_masked(X) or np.ma.is_masked(Y):
-> 5539 raise ValueError(
5540 'x and y arguments to pcolormesh cannot have '
5541 'non-finite values or be of type '
5542 'numpy.ma.core.MaskedArray with masked values')
5543 # safe_masked_invalid() returns an ndarray for dtypes other
5544 # than floating point.
5545 if isinstance(X, np.ma.core.MaskedArray):
ValueError: x and y arguments to pcolormesh cannot have non-finite values or be of type numpy.ma.core.MaskedArray with masked values

This error:
ValueError: x and y arguments to pcolormesh cannot have non-finite values or be of type numpy.ma.core.MaskedArray with masked values
is because there are NaN’s in the coordinate variables, as explained in the plotting with cartopy tutorial.
By following the instructions in that tutorial and the Spatial selection with tripolar ACCESS-OM2 grid notebook the coordinates can be fixed by replacing them with coordinates from the ice grid input file. It requires some work, the dimensions must be renamed to match, and coordinates converted from radians to degrees.
[27]:
ice_grid = xr.open_dataset('/g/data/ik11/inputs/access-om2/input_eee21b65/cice_025deg/grid.nc').rename({'ny': 'nj', 'nx': 'ni'})
ice_grid = ice_grid.pint.quantify()
ice_air_temp = ice_air_temp.assign_coords({'TLON': ice_grid.tlon.pint.to('degrees_E'), 'TLAT': ice_grid.tlat.pint.to('degrees_N')})
ice_air_temp
[27]:
<xarray.DataArray 'Tair_m' (time: 12, nj: 1080, ni: 1440)>
dask.array<concatenate, shape=(12, 1080, 1440), dtype=float32, chunksize=(1, 1080, 1440), chunktype=numpy.ndarray>
Coordinates:
* time (time) datetime64[ns] 1958-02-01 1958-03-01 ... 1959-01-01
TLON (nj, ni) float64 [degrees_east] -279.9 -279.6 -279.4 ... 80.0 80.0
TLAT (nj, ni) float64 [degrees_north] -81.08 -81.08 ... 65.13 65.03
Dimensions without coordinates: nj, ni
Attributes:
units: C
long_name: air temperature
cell_measures: area: tarea
cell_methods: time: mean
time_rep: averaged
ncfiles: ['/g/data/hh5/tmp/cosima/access-om2-025/025deg_jra55v13_i...Finally, the generic plotting routine works
[28]:
plot_global_temp_in_degrees_celcius(ice_air_temp)
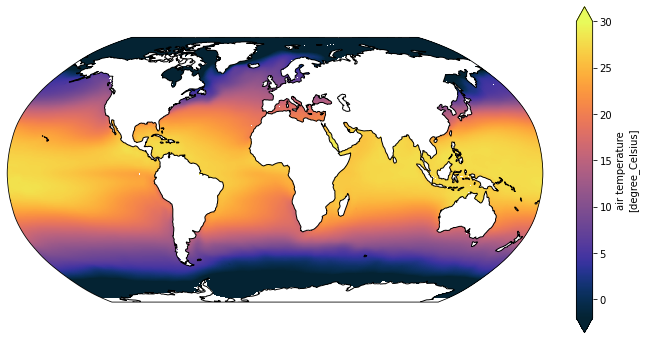
One more step is to modify the original routine to take the vertical range as an argument, so it is more generally useful:
[29]:
def plot_global_temp_in_degrees_celcius(da, vmin=-2, vmax=30):
# Take the time mean of da and plot a global temperature field in a Robinson projection
#
# Input DataArray (da) should be a 3D array of latitude, longitude and time.
da = da.pint.quantify().pint.to('C')
da_time_mean = da.cf.mean('time')
plt.figure(figsize=(12, 6))
ax = plt.axes(projection=ccrs.Robinson())
da_time_mean.cf.plot(ax=ax,
x='longitude', y='latitude',
transform=ccrs.PlateCarree(),
vmin=vmin, vmax=vmax, extend='both',
cmap=cm.cm.thermal)
ax.coastlines();
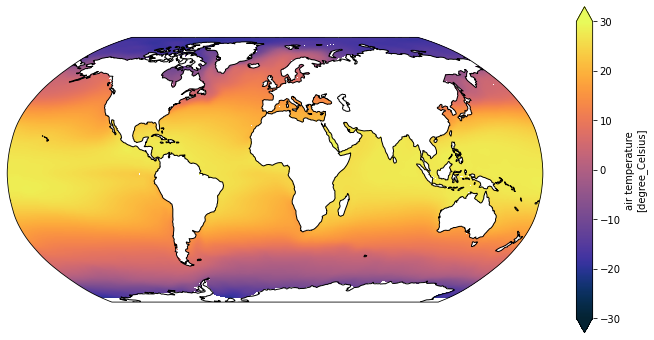
By specifying default values for the arguments it is completely backwards compatible, we have lost no functionality, but the ice air temperature can now be plotted with a range that better suits the range of the data
[30]:
plot_global_temp_in_degrees_celcius(ice_air_temp, vmin=-30)
Conclusion¶
Model specific code to take a time mean and plot the data was converted to a model agnostic function with no loss of functionality.
The same function can be used with a wide range CF compliant data.
In some cases the input data will need to be modified if it is not sufficiently compliant, or non-conforming in some way, such as the ice data above with NaN’s in the coordinate. It is better to modify the data to be more compliant and higher quality, and use generic tools, than have multiple code versions to account for the vagaries or problems with individual datasets.
Ideally those improvements can be incorporated into future versions of the data at source, in post-processing, or in some utility functions for transforming a class of non-conforming data.